The iglu package includes two single page reports - an ambulatory glucose profile (AGP), and an episode calculation report.
Ambulatory Glucose Profile (AGP)
The iglu package allows one to generate an Ambulatory Glucose Profile (AGP) report - see Johnson (2019) “Utilizing the ambulatory glucose profile to standardize and implement continuous glucose monitoring in clinical practice.”. Below is an example report for Subject 1, which includes information on data collection period, time spent in standardized glycemic ranges (cutoffs of 54, 70, 180 and 250 mg/dL) displayed as a stacked bar chart, glucose variability as measured by %CV, and visualization of quantiles of the glucose profile across days together with daily glucose views.
agp(example_data_1_subject)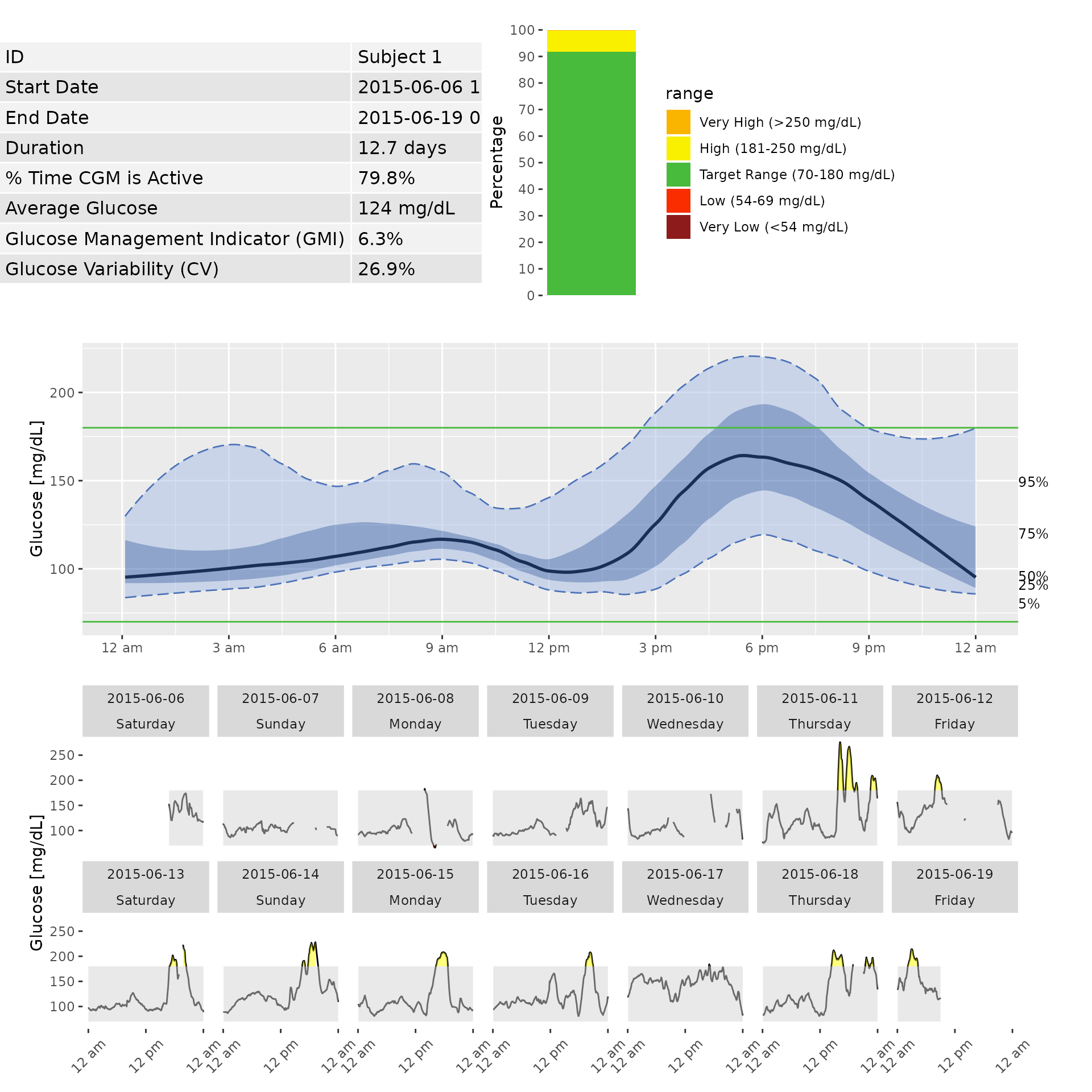
Episode Calculation
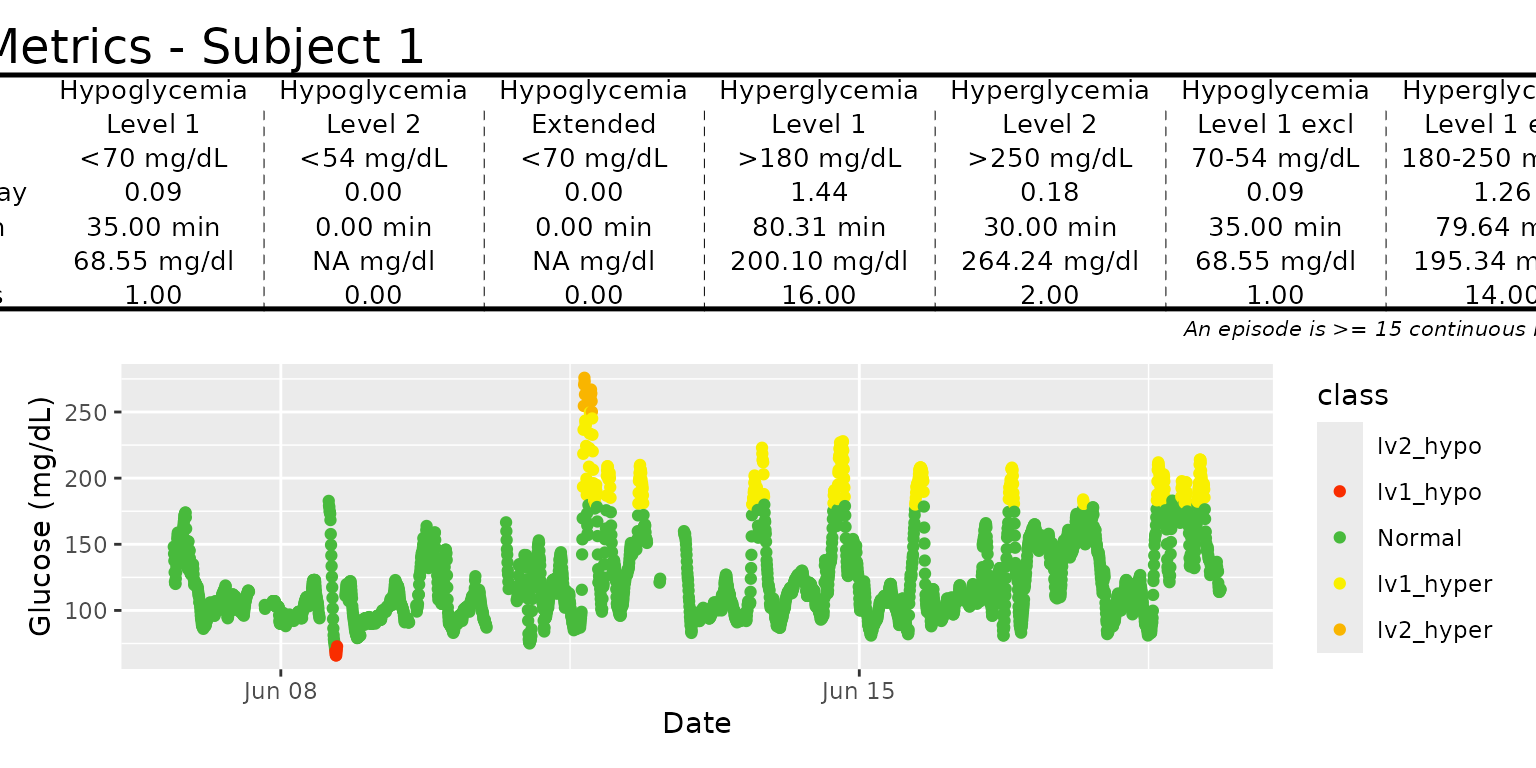
The episode_Calculation function measures the number of hypoglycemia and hyperglycemia episodes or events.
episode_calculation(example_data_1_subject,lv2_hypo = 70, lv1_hypo = 120, lv2_hyper = 180, dur_length = 15)
#> # A tibble: 7 × 7
#> id type level avg_ep_per_day avg_ep_duration avg_ep_gl total_episodes
#> <fct> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Subject 1 hypo lv1 3.33 257. 105. 37
#> 2 Subject 1 hypo lv2 0.0899 35 68.6 1
#> 3 Subject 1 hypo exten… 1.71 437. 101. 19
#> 4 Subject 1 hyper lv1 1.44 80.3 200. 16
#> 5 Subject 1 hyper lv2 1.44 80.3 200. 16
#> 6 Subject 1 hypo lv1_e… 3.24 262. 105. 36
#> 7 Subject 1 hyper lv1_e… 0 0 NA 0In this example, example_data_1_subject contains multiple days, and episode_calculation function calculate the episodes across all days.
Parameters
lv1_hypo, lv2_hypo, hy1_hyper, lv2_hyper
Users can set certain thresholds for the hypo and hyperglycemia by passing parameters, lv1_hypo, lv2_hypo, lv1_hyper, lv2_hyper. Level 2 indicates more severe states than level 1 so the threshold value for the lv2_hypo value should be lower than lv1_hypo value, and the threshold for the lv2_hyper value should higher than lv1_hyper value.
Epicalc_profile function
Visualization of the metrics produced by the
episode_calculation() function is done with the function
epicalc_profile(). This function takes the
episode_calculation() output and displays it as a tables of
the episode metrics as well as plots that visualizes the subject’s
episodes. This function is designed to work with one subject data at a
time, and the structure of the function output is shown below.
epicalc_profile(example_data_1_subject)